Introduction
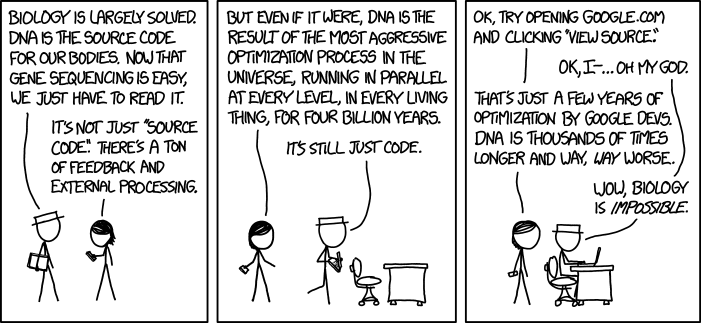
From early 2018 until late 2022 I worked as a software developer attached to a bioinformatics team. One of the things I saw the team around me doing was a lot of "scoring" of sequences, etc (imagine a lot of layman hand-waving here, obviously).
Playing on the "DNA is just code" nonsense trope, it became a bit of a running joke for me to suggest that they should try a scoring system based on a popular tile-based word game. Eventually, with a bit of spare time on my hands, I decided to actually write and publish a library that implemented this.
And so the Flounder Score was born.
No, it's not a serious library, but I also think it makes a serious point about people who think "it's just code".
Installation
The library can be installed from PyPI:
$ pip install flounder-score
Usage
See the module documentation for the detailed library documentation.
Getting the score
This function takes a string that is a sequence returns an integer value that is the flounder score for that sequence. Case is not important. IUAPC codes are taken into account.
Example:
from flounder import score
print(score("GTAC"))
7
Getting all the scores
This function takes a string that is a sequence, and returns a list of
tuples. Each tuple is the base at that position in the input string, along
with its individual score.
Example:
from flounder import scores
print(scores("gtac"))
[('g', 2), ('t', 1), ('a', 1), ('c', 3)]
Getting the score to the max
This function takes a string that is a sequence, and returns a "to the max" score. Case is not important. IUAPC codes are taken into account.
The difference with this scoring is that, for the bases, not only do they
score for their own score, they also
score for every IUAPC
code that is related to that
base.
Example:
from flounder import score_to_the_max
print(score_to_the_max("gtac"))
86
Getting all the scores to the max
This function is similar to scores, except it
returns a list of tuples where each tuple is the base at that position in
the input sequence, along with its individual
score_to_the_max.
Example:
from flounder import scores_to_the_max
print(scores_to_the_max("gtac"))
[('g', 19), ('t', 24), ('a', 20), ('c', 23)]
Getting the codon score
This function takes the input sequence, translates it into an amino acid sequence (using as many codons as it can find from the first position), and then scores that resulting sequence.
For example:
from flounder import codon_score
print(codon_score("AGACGCAGTCTT"))
4
Getting the codon scores
This function takes an input sequence, translates it into an amino acid sequence (using as many codons as it can find from the first position), and then returns a list of tuples where the value in the first position is the codon being scored and the second is the score for that codon.
For example:
from flounder import codon_scores
print(codon_scores("AGACGCAGTCTT"))
[('AGA', 1), ('CGC', 1), ('AGT', 1), ('CTT', 1)]